Article introduction
Prostate cancer (PCa), the second most common cancer in males, seriously affects the reproductive health and quality of life of patients. Because prostate cancer is regulated by a variety of genes, and there is currently no in-depth study on related mechanisms, the treatment effect is often not satisfactory. With the development of high-throughput technology, it is possible to study the genes and mechanisms of action of PCa-related molecular markers. At present, it has been reported that many star LncRNAs such as SChLAP1, HOTAIR, PCA3, PCAT1, NEAT1 and CTBP1-AS play an important role in PCa. So the question is, these LncRNAs are mostly located on the autosomes, but PCa can be used as a male-specific disease. Should the main genes regulating its function be located on the Y chromosome?
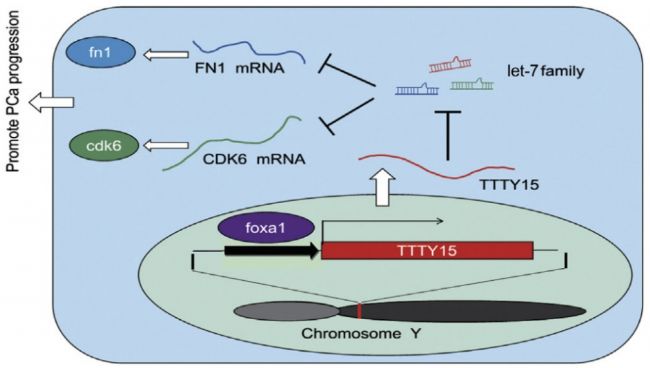
With this problem, the author of this article uses the LncRNA on the Y chromosome as an entry point to rapidly target a highly differentially expressed LncRNA in PCa by high-throughput screening technology, and confirmed it through a large number of clinical samples. It is indeed highly expressed in PCa patients. Finally, it was confirmed by molecular research that this process was regulated by miRNA-let-7, the target genes CDK6 and FN1, and the transcription factor foxa1. 

Article content
1. LncRNA-TTTY15 is highly expressed in prostate cancer tissues
This time, the research team conducted deep bioacoustic mining on the LncRNA part of the whole transcriptome sequencing (the cloud sequence organism can provide this service) data, and found that LncRNA-TTTY15 located on the Y chromosome is highly expressed in prostate cancer tissues. The results of the experiment were published again in the journal European Urology. Through LncRNA quantitative PCR , the authors in the sequencing sample (65:65), increased clinical samples (35:35) for large sample low-throughput validation, mining sequencing data from other ethnic prostate cancer patients in the TCGA database, and through larger clinical samples Transcriptome sequencing (145: 145) further confirmed the high expression of TTTY15 in prostate cancer tissues. At the same time, in situ hybridization experiments at the tissue level also confirmed its specific expression in cancer tissues.

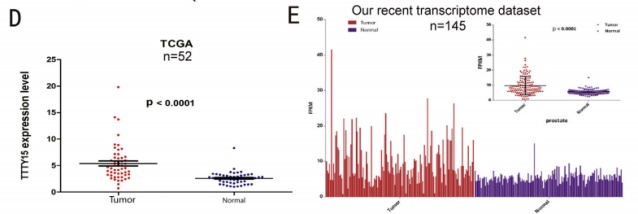
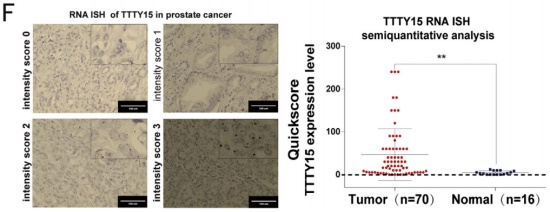
2. TTTY15 promotes prostate cancer cell proliferation and migration
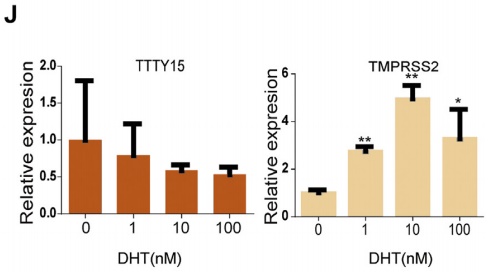
The authors selected a total of six cell lines, and found that TTTY15 is highly expressed in cancer cells by LncRNA quantitative PCR. Subsequently, the authors selected several highly expressed cell lines, and inhibited and overexpressed the TTTY15 gene, respectively, and found that the proliferation rate and number of cells were significantly affected. However, after overexpressing TTTY15 in an androgen-related cell line, the LncRNA was found to be insensitive to androgen, suggesting that its upstream factor may not be an androgen receptor.
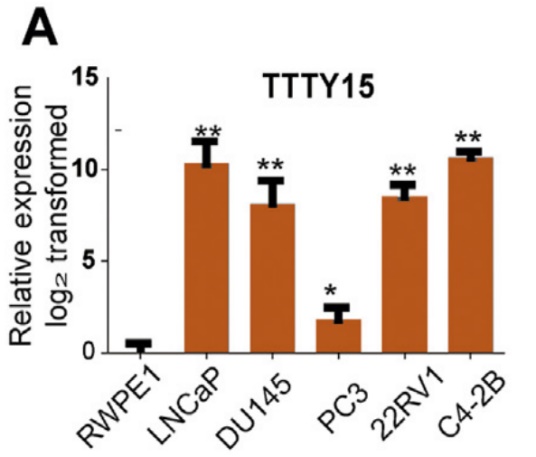
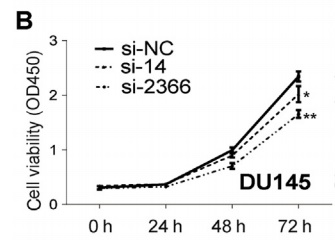
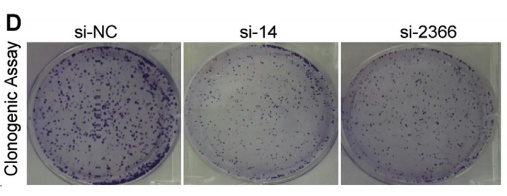
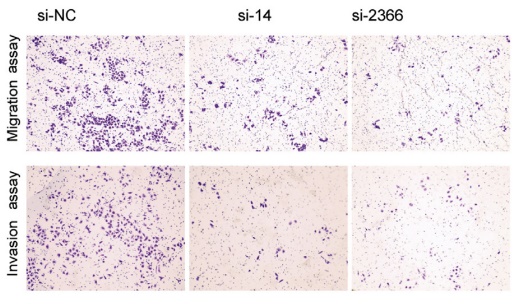
3. CRISPR/Cas9 inhibits the promotion of LncRNA TTTY15 on prostate cancer cells
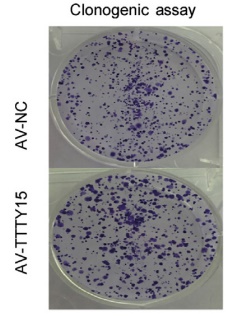
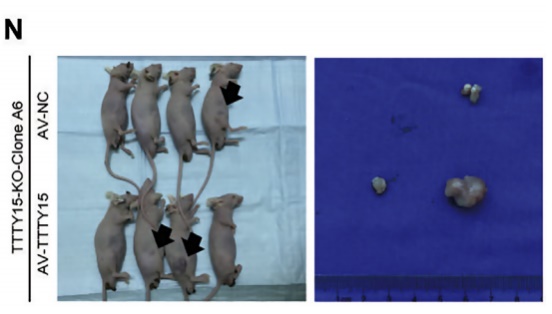
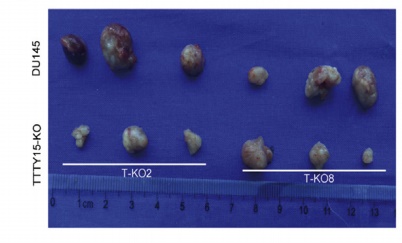
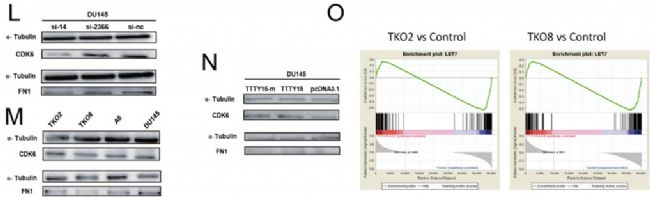
The authors of this article knocked out LncRNA TTTY15 by two CRISPR/Cas9 strategies, the first knocking out the entire TTTY15 fragment; the second is terminating TTTY15 transcription by knocking in the transcription terminator. Regardless of the method of knockout, both proliferation and migration were inhibited at the cellular level, and complementation experiments showed that inhibition could be restored. At the same time, after the authors transplanted knockdown cells into mice, the mice in the transplant group were smaller than the control group. Overall, the authors have fully demonstrated that TTTY15 promotes prostate cancer from both cell and animal dimensions.
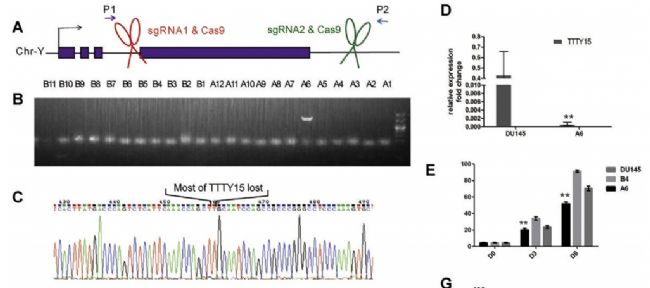
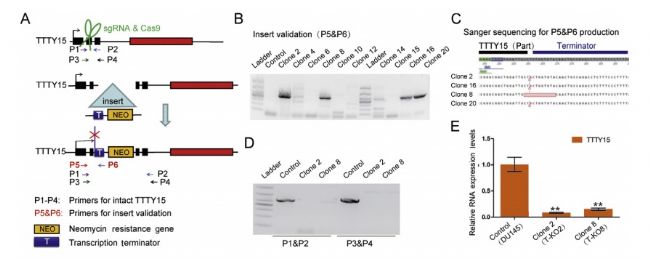
4. LncRNA TTTY15 plays a sponge mechanism by adsorbing miRNA
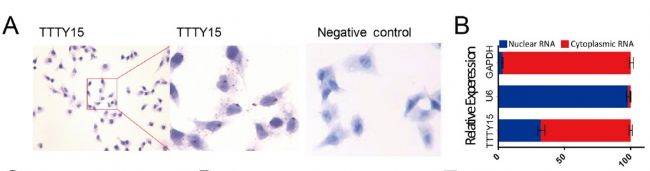
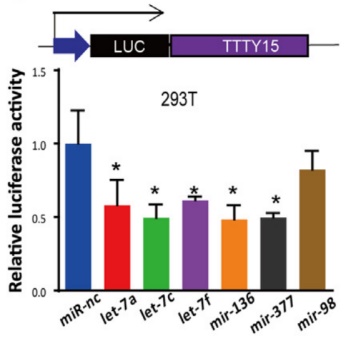
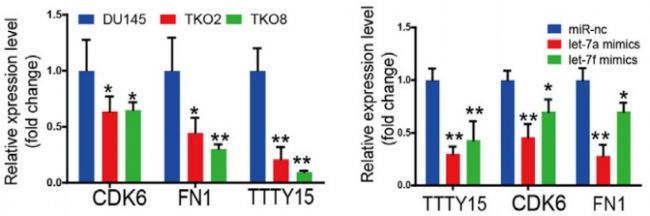
First, the authors confirmed by cell localization experiments that LncRNA TTTY15 is mainly distributed in the cytoplasm, which indicates that the LncRNA may be related to the sponge mechanism. Next, the possible binding of miRNAs was predicted by a joint analysis of miRDB and miRanda software. The dual luciferase reporter gene experiments showed that there were 5 miRNAs bound to it, and the expression of miRNA let-7 was negatively correlated with LncRNA by miRNA quantitative PCR . Next, the authors verified 22 target genes that have been reported to bind to let-7 in LncRNA knockdown and overexpressing cell lines by mRNA quantitative PCR , and screened two target genes co-expressed with LncRNA. They are CDK6 and FN1. At the RNA, protein level and biosignal GSEA predictions, the expression level of the target gene was confirmed to be inversely proportional to let-7.
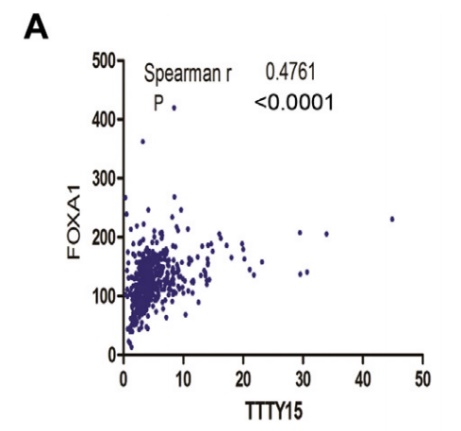
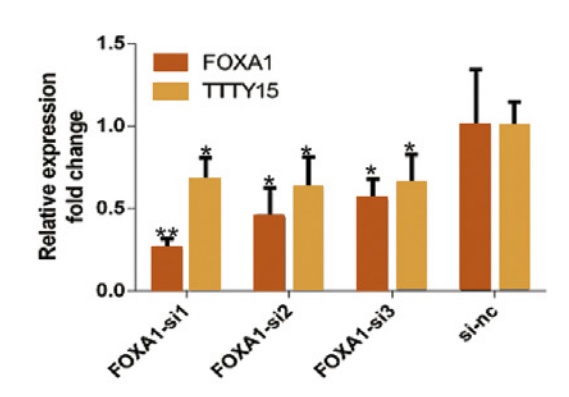
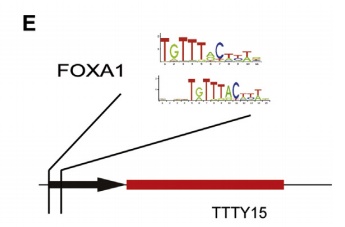
5. FOXA1 is the upstream transcription factor of LncRNA TTTY15
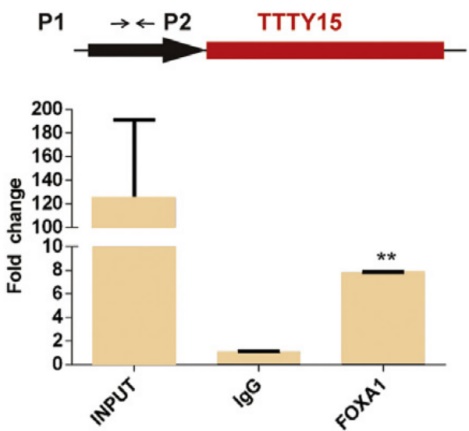
By prediction, the authors predicted many hypersensitive and epigenetic molecular marker binding sites on the LncRNA TTTY15 promoter. The TCGA database showed that only the expression levels of FOXA1 and FOXB13 were positively correlated with LncRNA. After knocking down the two transcription factors, the authors found that only FOXA1 inhibited the expression of LncRNA, and FOXA1 was also highly expressed in prostate cancer tissues. In the end, the authors confirmed that the two are directly related by ChIP sequencing (the cloud sequence organism can provide this service) .

to sum up
It is worth mentioning that Yunzo Bio has been very fortunate to participate in the transcriptome data mining of 130 cancer and paracancerous tissues of prostate cancer patients in this article, which provides the basic work for the discovery of LncRNA TTTY15.
Review the idea of ​​this article: Firstly, through the whole transcriptome sequencing data, we screened a LncRNA TTTY15 with the highest expression in prostate cancer tissue and located on the Y chromosome, and then combined with biosignal prediction, quantitative PCR, dual luciferase reporter gene and In experiments such as ChIP sequencing, the authors confirmed that LncRNA TTTY15 adsorbs let-7 through sponge action and promotes the expression of target genes CDK6 and FN1, thereby promoting the development of prostate cancer.
Full text link:
Https://(17)30720-0/fulltext
The cover image is taken from:
Https://
Cloud order related product recommendation:
Whole transcriptome sequencing
Circular RNA sequencing
LncRNA sequencing
ChIP sequencing
RNA pull down
RIP sequencing
Circular RNA quantitative PCR
LncRNA quantitative PCR
miRNA quantitative PCR
Quantitative PCR
Past review:
Multi-omics association analysis reveals new genetic changes in prostate cancer
Nature Subsidiary | circRNA_104075 sponge adsorbs miR-582-3p, which inhibits YAP expression, thereby promoting the development and progression of liver cancer
Nature | SMAD2/3 and TGF-β pathway synergistically affect transcription factor m6A RNA methylation regulates stem cell development
Nat. commun|Mettl3-mediated m6A RNA methylation regulates bone marrow mesenchymal stem cells and osteoporosis
Nature|m6A RNA methylation recognition protein YTHDF1 is involved in memory formation
No, Da Niu tells you how to study lncRNA methylation
RNA methylation enters the ultra-micro era
More than 10 points of m6A RNA methylation sequencing article--Cloud order bio-assisted
RNA methylation in 20 points, coexistence of heat and strength
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - non-coding RNA articles
Cloud Sequence Bio's Latest "RNA Methylation" Research Summary - Arabidopsis
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - virus articles
Plant Cell: Arabidopsis found a new m6A RNA demethylation modifying enzyme
Cloud order customer 12 points top article, teach you how to use RIP sequencing to play the molecular mechanism!
2018 Nature Magazine's Breakthrough Research Results--m5C RNA Methylation Cloud Sequence Creature
Nature Breakthrough Research - RNA Methylation Newly Modified m1A Cloud Sequence Creature
Nature Heavy: m6A RNA methylation is involved in the key aspects of hematopoietic stem cell development!
Cancer Cell: Professor He Chuan discovers important functions of m6A RNA methylation
Xiaobai must see! Summary of methods for identification of overall level of RNA methylation
Hepatology: m6A RNA methylase METTL3 promotes the development of liver cancer
Yunxu Bio exclusive m5C RNA methylation sequencing
Nature article reveals multiple functions of RNA methylation
Filling the gap of epigenetic modification: a new mechanism for RNA methylation regulation gene export
Shanghai Yunxu Biological Technology Co., Ltd.
Shanghai Cloud-seq Biotech Co., Ltd.
Address: 3rd Floor, Building 20, No. 518, Zhangzhu Road, Songjiang District, Shanghai 
Telephone Fax Website:
mailbox:
Spinal Fixation System,Vertebral Arch Pedicle Screw,Monoaxial Pedicle Screw,Spinal Screw Shaft
Changzhou Ziying Metal Products Co., Ltd , https://www.ziyingmetal.com
