Science: the fork to the heart
March 13, 2018 Source: Biological Exploration of: Tierna
Window._bd_share_config={ "common":{ "bdSnsKey":{ },"bdText":"","bdMini":"2","bdMiniList":false,"bdPic":"","bdStyle":" 0","bdSize":"16"},"share":{ }};with(document)0[(getElementsByTagName('head')[0]||body).appendChild(createElement('script')) .src='http://bdimg.share.baidu.com/static/api/js/share.js?v=89860593.js?cdnversion='+~(-new Date()/36e5)];Congenital heart disease is one of the most common birth abnormalities and usually affects specific areas of the heart or heart cell type, and understanding the early cardiac lineage diversity is critical to revealing the origin of congenital heart disease. In addition, the directed differentiation of pluripotent stem cells into specific cardiac lineages is a key step in Drug Testing and regenerative therapy for heart disease.
According to a recent Science report, the researchers used single-cell RNA sequencing to generate two high-resolution snapshots of gene expression in the neonatal cardiovascular germ layer of early mouse embryos. This finding illustrates the transcriptional complexity of the anterior endoderm and reveals how different lineages that act on the heart during embryogenesis are produced.

Image source: Network
1 , the regulation mechanism of cell fate selection
Understanding the regulatory mechanisms that drive cell fate selection during pedigree diversification is a core goal of developmental biology. By dividing the transcriptional content of many independent cells into different places, single-cell transcripts can predict a revolution in our grasp—how ​​cell fate decisions occur. (See below). This technology can change our understanding of biological complexity and help us discover new cell types and regulatory mechanisms in the development of homeostasis and disease.
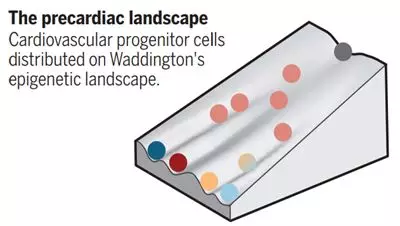
Image source: science (DOI: 10.1126/science.aat0230 the same)
2 , the formation of new heart
During embryonic development, cardiac progenitor cells transiently express the transcription factor mesodermal protein 1 (Mesp1) in gastrula. The neonatal heart mesoderm migrates to the lateral region of the embryo where the progenitor cells of the first cardiac region produce early cardiac tubes. Subsequently, the late differentiation into pluripotent progenitor cells of the second heart contributes to the growth of the heart from the adjacent pharyngeal mesoderm. The second heart cell produces the veins and arteries of the heart, which is a hotspot of congenital heart defects. The first and second heart fields are separated before the Mesp1 is expressed.
3. Single-cell RNA sequencing to determine the diversity of cardiac cell lineages
Single-cell RNA sequencing has recently provided insight into the degree of transcriptional diversity of cell types in different regions and developmental centers, providing valuable resources for discovering new pathways and genes associated with heart disease. Lescroart et al. focused early on the cells expressed by mespl, and when the gene was expressed on the first and second cardiac field progenitors, these new data were completely consistent with those produced by pre-jejunal cells and later mesoderm. Developmental continuum between single cell transcriptomes. Using a clustering algorithm to display the gene expression topology in these combined data sets, the authors identified different progenitor subpopulations of early lineage diversification. These cells include cardiomyocytes and endothelial progenitor cells (possibly the first heart-derived cells), as well as cells with the pharyngeal mesoderm genetic markers (defined as the posterior and anterior clusters of the second cardiac cells), the latter including the head Muscle progenitor cells. In the later stages of development, the posterior and anterior second heart cells act on the veins and arterial arches of the heart, respectively. In addition, the researchers also found that different progenitor cell clusters appear at the edge of the core of molecular heterogeneous cardiovascular progenitor cells.

Distinct progenitor cell clusters emerge at the edge of a transcriptionally heterogeneous core of nascent cardiovascular mesoderm
4 , research progress
Lescroart et al. calculated the trajectory of time to track the progression of pluripotent ectoderm and different progenitor cell states on the transcriptional profile. The results of the analysis showed that the anterior and posterior second cardiac field clusters were different from the normal progenitor cells expressing the genes enriched in the two lineages in the central nucleus. This multilineage priming was observed in Mesp1-labeled myocardium and pharyngeal muscle progenitors in the gut of the basal chord, indicating that this is a conserved feature of the separation of fate of the pharyngeal mesoderm cells. Cells within the core population may thus constitute a transitional state between different trajectories, where extrinsic signal events affect pedigree results. In the case of genetic or environmental perturbations, the transcriptional heterogeneity of the core population will lead to developmental plasticity and robustness, which may lead to compensatory mechanisms and phenotypic variation in patients with congenital heart defects.
Excavating data sets generated by Lescroart et al. helps to systematically define extrinsic and intrinsic regulators that control early mesodermal sequential fate decisions. For example, the authors converted Notch signaling into Mesp1, an early enrichment of endothelial cells and myocardial trajectories. Regulatory pathways. In addition, transcriptome analysis of single cells from mutant embryos revealed that Mesp1 itself controls the transition from pluripotency to progenitor cell size.
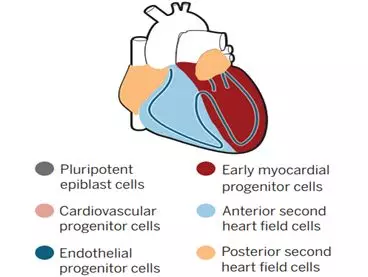
Distinct progenitor cell populations give rise to diferent regions of the heart and cardiac cell types
5 , the challenges encountered
A major challenge in single-cell transcriptomics analysis is mapping cells to the tissue of origin. Using fluorescence in situ hybridization to track selected genes, the authors found that myocardial and endocardial progenitor cells can be spatially resolved in the nascent mesoderm or in the anterior and posterior regions of the larynx.
A recently developed method for mapping individual cells after RNA sequencing utilizes a high-resolution reference map set generated by in situ hybridization of a series of landmark genes or spatial transcription methods, such as single embryo spatial RNA sequencing. The latter technique has been developed in a phase that overlaps with the work of Lesroart et al. and should allow topology mapping of this new data set. This will allow the identification of primordial cells on different trajectories and clarify our understanding of how future regions of heart and heart cell types form patterns in progenitor cell populations.
Another challenge in this high-throughput data set is the ability to screen out valid data from countless experiments, and analysis of additional cells and time points may interfere with our perception of progenitor cell structure. Combining the single-cell transcriptome approach with the single-cell epigenome dataset and lineage provides a fine-grained map of the dynamic cardiovascular progenitor landscape, guiding us to the selection mechanism that drives the fate of cardiac progenitors in development and disease. System exploration.
References: 1) Diverging roads to the heart
PCR ( Polymerase Chain Reaction)
It provides a way to make more copies of a portion of DNA.
We need to have PCR ingredients that include DNA portion, Buffer solution, Primer+DNA (tap polymerase), and Nucleotides during the preparation step.
In the step of PCR sequence, there are three steps in this section.
Step 1 Denaturation
The addition of heat is needed to separate the two strands of the DNA molecule.
Step 2 Annealing
Temperature, is going to be cool in this step, should allow the primers to bind to the specific segments.
Step 3 DNA Synthesis
To make more copies of the DNA.
Those three steps above keep repeating to make enough copies of the DNA fragments. It is the principle of how PCR machine works.
SARS-Cov-2, One test type that uses a sample from a nose or throat swab in a 'PCR test'. The goal is to make enough copies of the viral cDNA in order for it to be detectable.
Superyears General 2215 RT-qPCRadopts all-in-one with an industrial art design, combining the Analytical Instrument, Large touch screen, and Computer. General 2215 RT-qPCR is equipped with the proven thermal-cooled electric technology, which can recognize the speed rate of temperature control faster and obtain high-quality testing results within 40 minutes. To ensure the sensitivity and stability of the testing a design with traditional beam path with a susceptible CMOS image sensor is used. In addition, General 2215 supports multiple PCR analysis and High-resolution Melt Curve (HRM) analysis, compatible with the mainstream reagent kit in the market. It will be the best assistant and your preferred RT-qPCR.
Rt-Pcr For Sars-Cov-2 Testing,Real Time Pcr Machine Kary Mullis,Pcr Machine Covid-19 Nucleic Acid,Qpcr Machine Covid-19 Omicron Detection
Nanjing Superyears Gene Technology Co., Ltd. , https://www.superyearsglobal.com
