RNA methylation is currently the hotspot of natural projects in the applicant country, and it is the only field that can send dozens of nature and cell-level high scores in just 3 months. Recent RNA methylation research has caused researchers' research boom. Since mRNA is involved in protein coding, most of the previous studies have focused on mRNA methylation (see the review of the cloud sequence classroom for details). However, many studies have shown that non-coding RNAs that undergo m6A methylation play a key role in gene regulation, stem cell differentiation, cancer cell proliferation, tissue growth and development, etc., so research on non-coding RNA methylation is becoming more and more hot. Today, let us talk about m6A RNA methylation regulating star members in the non-coding RNA family: miRNA, LncRNA and circRNA.
1.Nature: m6A regulates the recognition of pri-miRNA
Impact factor: 40.10
The research team of Rockefeller University in the United States performed m6A RNA methylation sequencing on human breast cancer cells. High-throughput data statistics showed that there are many METTL3 motifs on the pri-miRNA targeted by m6A. Therefore, a preliminary investigation was conducted on whether METTL3 participates in the m6A methylation modification of pri-miRNA. After knockdown, overexpression, and cell localization experiments, it was confirmed that METTL3 binds to pri-miRNA. Further, by Clip sequencing, it was shown that METTL3 and pri-miRNA can be directly combined. This article is full of dry goods, and also details the mechanism of action of the two.
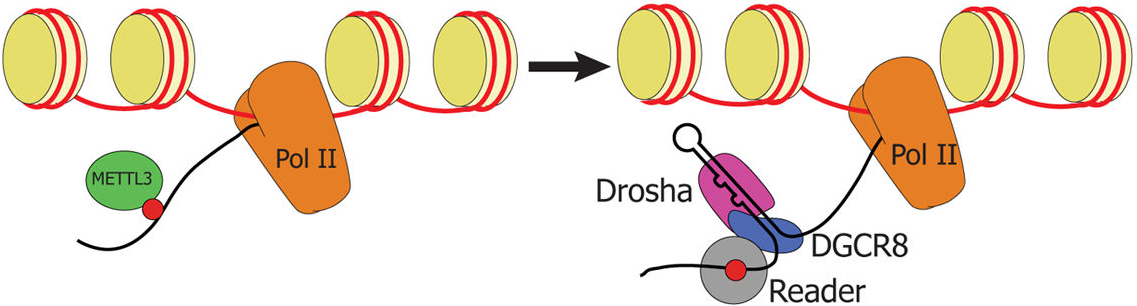
Figure 1. m6A methylation regulates pri-miRNA
2.Nature: m6A of LncRNA (MALAT1) regulates pre-mRNA synthesis
Impact factor: 40.10
The University of Chicago research team first reported a LncRNA (MALAT1), which is associated with human lung cancer, and has an m6A modification on its stem-loop structure. Through RNA pull down experiments , the authors found that the m6A methylation modification of this site enhanced the binding ability to hnRNP C protein. It was also confirmed that methyltransferases METTL3 and METTL14 are also involved in this process. Teachers who want to study single-site RNA methylation can refer to this article.
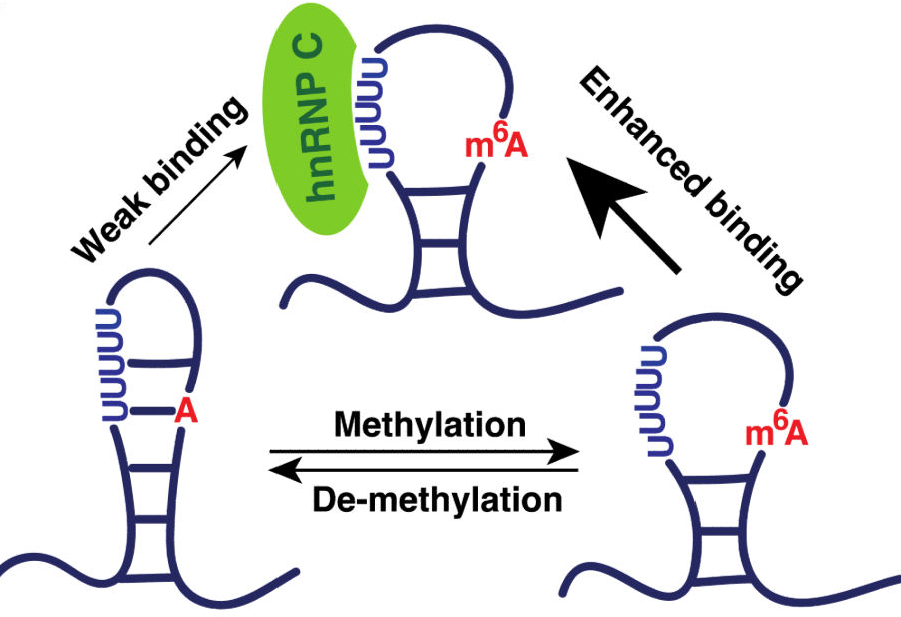
Figure 2. Modification of m6A methylation modification on MALAT1 in combination with hnRNP C
3. Cancer Cell: m6A methylation regulates LncRNA FOXM1-AS promotes cancer cell proliferation
Impact factor: 27.41
The authors analyzed multiple cancer-related databases and predicted that the ALKBH5 gene is associated with a poor prognosis for malignant glioma. After the functional experiment, the function was also confirmed. Subsequently, the authors used the knockdown of the ALKBH5 gene as a sample, performed m6A RNA methylation sequencing and transcriptome sequencing, and quickly found the target gene FOXM1. LncRNA (FOXM1-AS) was anchored from the position and expression of the sequence, and it was confirmed by RNA pull down and complementation experiments that the two could be directly combined. High-throughput statistical results indicated that the 3'-end FOXM1-AS gene was methylated. RIP sequencing showed that ALKBH5 was directly proportional to the methylation level of FOXM1-AS. In this paper, the existing transcriptome data is used to determine the function of related genes, and then the mechanism research is carried out. This research idea is worthy of reference.
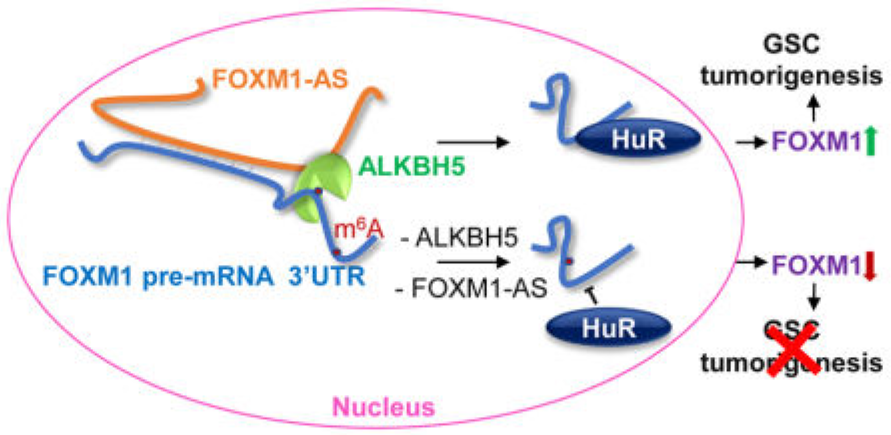
Figure 3. Methylation modification on LncRNA FOXM1-AS regulates cancer cell proliferation
4. Nucleic Acids Research: m6A methylation regulates lincRNA 1281 affects mouse embryonic stem cell differentiation
Impact factor: 10.16
The lincRNA is called a large intergenic non-coding RNA. First, the author first determined that linc1281 plays a guiding role in the proliferation of mouse embryonic stem cells. Subsequently, it was detected that the linc1281 gene was highly methylated, and there were three RRACU methylation motifs on the last exon. After overexpression, they did not directly cause the change of linc1281 gene expression. Sometimes no change is also a result, but fortunately the author did not give up, and finally found that linc1281 can regulate the ceRNA mechanism.
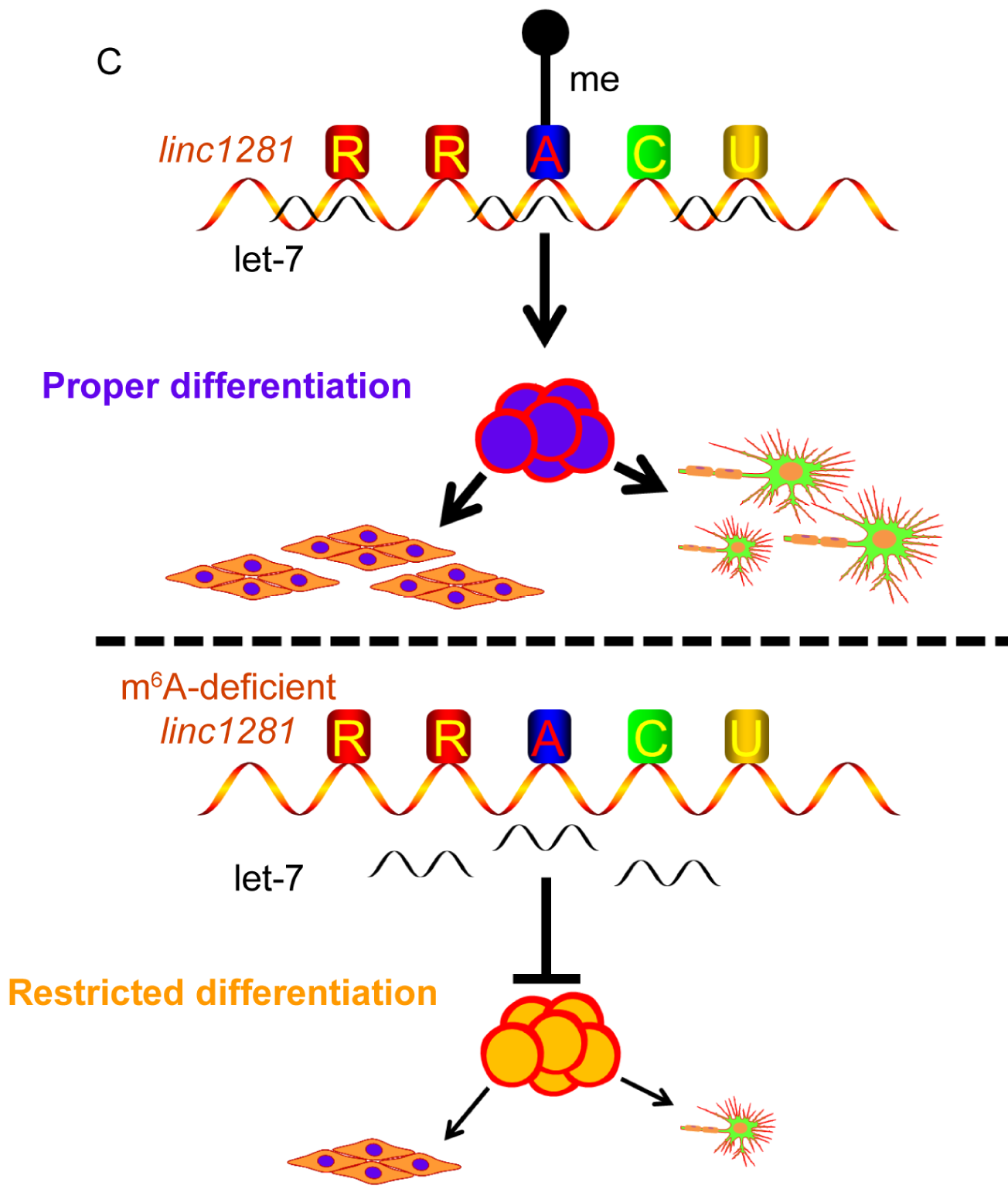
Figure 4. M6A methylation modification on lincRNA 1281 regulates mouse embryonic stem cell differentiation
5.Cell Research: m6A regulates circRNA-encoded proteins
Impact factor: 15.6
When it comes to the high-scoring article of circRNA, there is one that can't be circumvented. He is what we are going to say now. He came from the team of Wang Zefeng of Zhejiang University and confirmed for the first time that m6A methylated circRNA is involved in the protein coding process. Let's take a look at how Wang has studied the process. With RIP sequencing , the authors found that m6A methylation modification is more extensive in circRNA than mRNA. The author also found an unacceptable thing. Previously, it was confirmed that after transient heat treatment of cells, m6A methylation modification in intracellular mRNA can affect protein translation. So along this line of thought, the author wonders if the methylation of circular RNA will affect protein translation. As soon as the results were verified, it was found to have an impact. After that, the author has done the verification of the methylation-related enzyme mechanism according to the previous ideas. Xiaobian does not say much, I believe that the cloud powder can hear the ears.
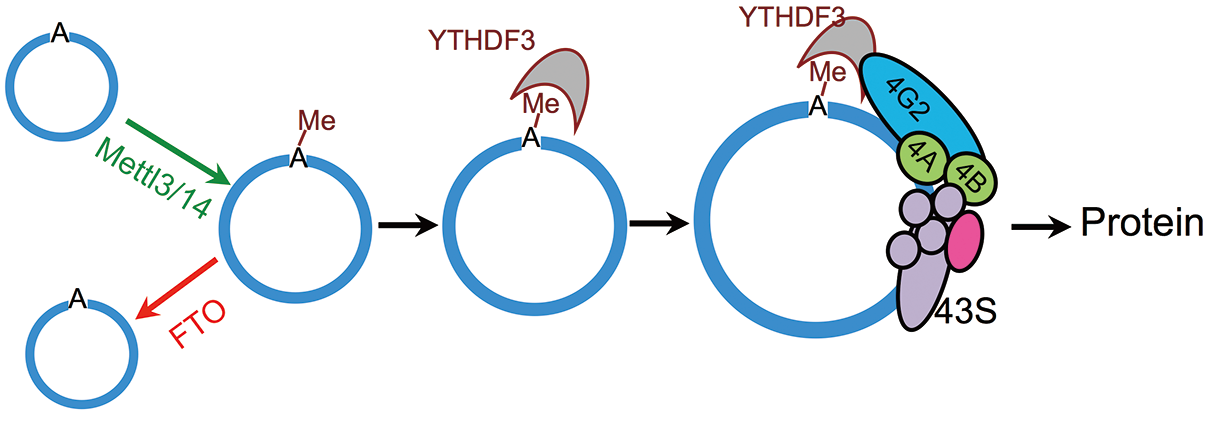
Figure 5. m6A regulates the mechanism of circRNA-encoded proteins
6.Cell reports: mtRNA overall m6A methylation profile
Impact factor: 8.28
The Harvard Medical School research team found that m6A modifications are widely present in circRNA in embryonic stem cells and cancer cells, and that the same circRNA is differentially distributed in different cells. Functional studies indicated that the m6A methylases METTL3, YTHDF1, and 2 are involved in the circRNA m6A modification process. If you want to know about circRNA, check out the comparative analysis of mRNA and m6A methylation in circRNA.
Figure 6. Differential expression of circRNA and genes in different tissues
Cloud sequence bio recommended:
- m6ARNA methylation sequencing
- m5CRNA methylation sequencing
- m1ARNA methylation sequencing
- RIP sequencing
Past review
- The latest cloud sequence biology m6A "RNA methylation" research summary - virus articles
- The latest cloud sequence biology m6A "RNA methylation" research summary - cancer articles
- The first domestic 10 points or more m6A RNA methylation sequencing article - cloud sequence bio-help!
- Yunxu Bio exclusive M5C RNA methylation sequencing
- 2018 National Nature Research Hotspot 2: In-depth analysis of RNA methylation research
- How to use RNA methylation sequencing data to publish 10 articles
Shanghai Yunxu Biological Technology Co., Ltd.
Shanghai Cloud-seq Biotech Co.,Ltd 
Address: Lane 1066, Qinzhou North Road, Caohejing High-tech Development Zone, Shanghai
phone
Website:
mailbox:
Insulin Syringes Needle,Disable Syringe,Monoject Syringe,10 Ml Syringe
FOSHAN PHARMA CO., LTD. , https://www.foshanmedicine.com
