Confirmation Analysis of Structures of Leachable Impurities in Pharmaceutical Packaging Materials by Gas Chromatography-Orbitrap Mass Spectrometry
Key words
Extractables and leachables, Q Exactive GC, Orbitrap mass spectrometer, differential analysis, unknown identification, container containment systems, pharmaceuticals.
Foreword
Plastic, polymer and other pharmaceutical product-specific packaging materials can release potentially toxic chemical impurities. The detection of such substances is not only a hot spot in the pharmaceutical industry, but also a serious challenge for relevant analysts. In general, the main purpose of extractables and leachables (E/L) research is to qualitatively confirm, quantify, and minimize the amount of any contaminants that may migrate from the packaging material to the final product or drug. . "Extractables" refers to chemicals in a closed container system that can be extracted into a solvent under laboratory accelerated conditions. Among them, laboratory acceleration conditions include temperature rise and strong solvent, and the purpose of acceleration is to achieve maximum extraction without avoiding material degradation or variation. "Leachables" is defined as a chemical that can migrate from a package to a pharmaceutical product during the product's shelf life.
The potential and actual impacts on product users are summarized as follows:
• Extractables = Possible effects
• Leachables = actual impact
Test sample body of two types of impurities:
• extractables = container material
• Leachable = final product
Extractables research reduces risk of use primarily by rapidly identifying potential toxic leachables and selecting different packaging materials. Generally, for most pharmaceutical dosage forms, any material that is in direct contact with the active pharmaceutical ingredient (API) should be analyzed for extractables and leachables; in some cases, secondary or tertiary packaging ( For example, the label) also needs to be analyzed. Leachables can be introduced throughout the drug container containment system and throughout the production process. In addition, the drug can react with the packaging material to produce leachable impurities that can continue to occur during drug storage. 1 The experimenter completed the extractables comparison experiment by exposing the packaging materials to extreme solvents, pH and temperature conditions to observe changes in the harsh environment. At present, the detection and identification of all compounds is a very difficult task, and analysts need to apply various analytical techniques to ensure the accuracy and comprehensiveness of the substance confirmation.
Gas chromatography-mass spectrometry (GC-MS) is widely used in extractables research. Its application advantages are mainly reflected in good chromatographic resolution, reproducibility, peak capacity and, above all, the rich business that can be used to identify matches. Spectrum Gallery. At the same time, the packaging products contain a large amount of volatile and semi-volatile components, which is the main reason why this technology is suitable for such analysis. This test uses a new generation of GC-MS systems with ultra-high resolution and mass-accurate mass spectrometry systems to detect and characterize compounds in polymer gaskets (ring seals) used in packaging containment systems and sealing products. This lab is designed to demonstrate the complete workflow for qualitative analysis of chemical constituents in a ring seal. The focus of this process is to perform a target-free monitoring of the sample by a full-scale high-resolution mass spectrometry scan, and to obtain the exact mass of the compound with the advantage of ultra-high resolution. Mass spectrometry resolving power plays an important role in accurately estimating compound element composition, analytical structure, distinguishing co-elution and isobaric compounds. Fast scanning speed, high sensitivity and wide linear range facilitate simultaneous detection of high and low abundance compounds. These instrumental features, combined with a unique software system that performs automated deconvolution algorithms and sample comparisons, form a powerful platform for structural analysis of complex compounds.
Experimental condition
Sample and pretreatment
A total of four ring seal samples were tested for this leaching impurity study; A – red seal, B – brown seal, C – white seal, D – black seal. The experimental personnel performed accelerated dissolution experiments in accordance with the requirements of the BioPhorum Operation Group (BPOG). The samples were cut into 20 mm sections and immersed in sealed screw cap vials containing 10 mL of 100% ethanol, 50% ethanol, water for injection (WFI) and 5M sodium chloride (NaCl) and allowed to stand at 40 °C for 30 days. The corresponding blank solvent samples for chromatographic comparisons were processed using the same method. An aliquot of each sample extract was placed in a GC vial for analysis. All liquid samples were subjected to liquid-liquid extraction with dichloromethane before entering the GC analysis.
Instrument parameters and method settings
All experiments were tested using a Thermo ScientificTM Q ExactiveTM GC OrbitrapTM GC-MS/MS system. The injection procedure was performed using a Thermo ScientificTM TriPlusTM RSH autosampler with a Thermo ScientificTM TRACETM 1310 gas chromatograph and a Thermo ScientificTM TraceGOLDTM TG-5SilMS column with a 10 m precolumn (30 m × 0.25 mm ID) × 0.25 μm, P/N 26096-1425) for chromatographic separation. Other instrument parameters are shown in the table below.

The Q Exactive GC system uses the EI Level 1 full scan mode with a mass resolution set to 60,000 (FWHM, m/z 200). In addition, positive chemical ionization (PCI) analysis is performed using methane as a reactive gas to obtain the identification of unknown components.
The molecular ion information you want.
data processing
Data acquisition using Thermo ScientificTM TraceFinderTM software. The software package is a single platform control, which can realize instrument control, instrument method establishment, optimization, and the overall workflow of data processing guided by qualitative and quantitative analysis. The TraceFinder software also features deconvolution and spectral alignment of high resolution mass spectra.
Results and discussion
The main purpose of this test was to analyze the solvent leachables in the ring seal using a non-target, first-mass full-scan data acquisition mode. By comparing the two samples with the control sample (blank ethanol), the chromatographic peaks unique to the ring seal extract were found and identified. In addition, the experimenter expects to be able to quickly identify compounds using the exact mass and spectral matching information of the sample compounds. To accomplish the above goals, we designed and implemented a complete workflow. The details of this workflow are summarized in Figure 1.
Separation of unique chemical components
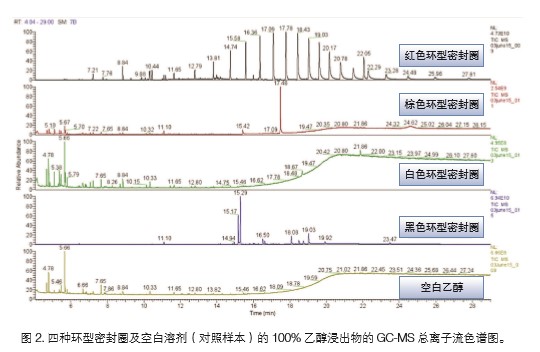
A full-scale mass spectrometry analysis was performed on all samples, and total ion chromatograms (TICs) are shown in Figure 2. The Q Exactive GC system enables accurate mass-to-charge ratio detection over a wide dynamic range, so that regardless of compound concentration, it can be detected indiscriminately without sacrificing mass accuracy. This ability enhances the confidence of identifying unknown compounds in complex samples. The first step in data analysis is to find peaks that are unique or significantly increased in each sample by comparison with blank samples. Although individual peaks can be seen in the TICs, in order to fully characterize the compounds in the test sample without missing any potentially toxic compounds, it is still necessary to perform an extraction analysis of all peaks in the data.
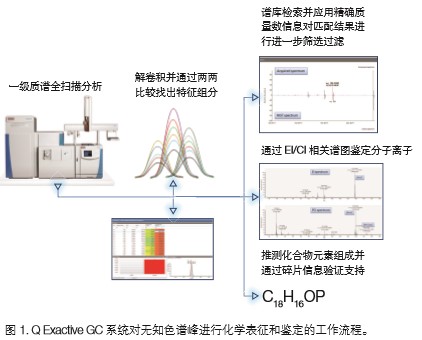
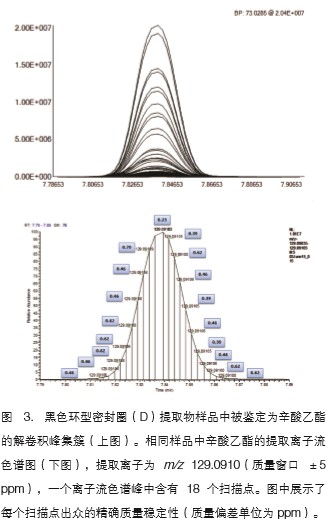
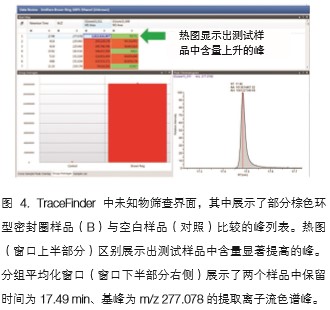
The TraceFinder software was used to perform a two-to-two comparison between the test sample and the blank sample to complete the data characterization. The software first performs accurate mass deconvolution analysis to find all ion peaks with a signal-to-noise ratio higher than 30:1, and removes the peaks in the mass spectrum of the same retention time as much as possible to maximize the compound ion peak for standard Library matching. Figure 3 shows the deconvoluted peak and the scan point of the entire chromatographic peak and the exact mass deviation of each base peak (m/z 129.0910). The extracted peak list is compared with the blank sample to classify the peak components characteristic of the test sample. The TraceFinder software also quickly finds elevated levels of components in the test sample in the form of a heat map (Figure 4). For example, a peak with a retention time of 17.49 min and a base peak of m/z 277.07800 is significantly elevated in the brown ring seal.
Identify compounds by library matching
After finding the specific chromatographic peak, we need to identify the compound. Applying TraceFinder software automates the qualification step (Figure 5). First, the deconvoluted mass spectrum of the compound is searched and matched in an existing commercial standard spectrum library (for example, NIST 2014), and all search results will be combined with the search for positive correlation (SI) scores and high resolution filter (HRF) values. Sort by comprehensive scores. Where the HRF value represents the percentage of the exact mass-to-charge ratio of the fragment peaks in the mass spectrum that corresponds to the corresponding fragment element composition in the standard library.

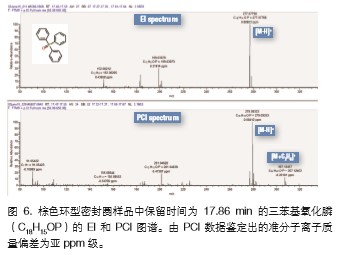
The rapid structural confirmation of unknown compounds can be achieved by combining accurate mass matching results and interpretation analysis of the composition of the fragment ions in the spectrum. For ion current peaks with a retention time of 17.49 min, the highest ratio of library matching is triphenylphosphine oxide, and the exact proportion of ions in the spectrum is 98.8%. The exact mass of the fragment ion is less than 1 ppm from the elemental mass of the corresponding fragment in the standard spectrum, which greatly increases the confidence in the identification results. The base peak m/z 277.07790 in the sample spectrum corresponds to the [MH]+ ion in the standard spectrum, and the exact mass of the ion is calculated by its elemental composition C18H14OP as m/z 277.07768, which deviates from the measured spectrum by 0.8 ppm. If the search results are sorted by only the traditional search index, many compound results (>700) with the same good match can be retrieved. Since the exact mass of the ion does not match the measured spectrum, these results are automatically filtered in this analysis.
In addition, we can further confirm the structural identification accuracy of triphenylphosphine oxide by evaluating the corresponding PCI spectra (Figure 6). In the PCI data, common additive ions such as [M+H]+ or [M+C2H5]+ can be found to estimate the elemental composition of the parent ion to evaluate whether the corresponding accurate mass deviation is within the ideal deviation (<1). Ppm). In the PCI spectrum of triphenylphosphine oxide, we found the associated adduct ions [M+H]+ (mass deviation 0 ppm) and [M+C2H5]+ (0.2 ppm) with ultra-high quality accuracy. And [M+C3H5]+ (0.4 ppm).
Identification of compound structures without spectral library matching results
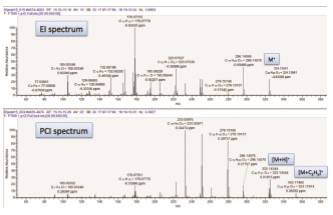
In the case where the EI spectrum cannot be retrieved in the standard library, we can estimate the elemental composition of the parent ion through the accurate mass information in the PCI spectrum. Good quality accuracy can effectively reduce the number of speculative results of elemental composition, which is especially necessary to improve the structural identification accuracy of compounds. An ion current peak with a retention time of 15.17 min in the black ring seal sample did not find the ideal match in the NIST library. All relevant search results were excluded due to discrepancies in accurate mass data. The excimer ions are estimated by the additive ion information in the PCI data (Fig. 7). The map shows that the ion m/z 325.14344 is [M+H]+(0 ppm) and the m/z 353.17483 is [M+C2H5]+ (0.3 ppm). The above ions indicate that the m/z 324.13541 in the EI spectrum should be a molecular ion peak. The elemental composition of the parent ion can be inferred by the molecular ion. This step analysis is a key step in the identification of compound structures. At this point, good mass accuracy reduces the number of possible elemental compositions. For example, if the mass accuracy tolerance window is set to 10 ppm, in the carbon atom (1-30), hydrogen atom (1-60), nitrogen atom (1-5), oxygen atom (1-5), phosphorus atom (1- There are 16 possible elemental compositions within the range of 5) and sulfur atoms (1-2). In contrast, if the mass accuracy tolerance window is set to 1 ppm, there is only one possible element component: C20H20O4. Only by controlling the quality accuracy to a small range can the number of elements that need further inspection be effectively reduced, and the credibility of the analysis results can be improved. The above-mentioned element composition estimation results are confirmed by the accurate mass information of [M+H]+ and [M+C2H5]+ adduct ions in the PCI spectrum. Except for this ion peak with a retention time of 15.17 min, the ion peaks with a retention time of 15.29 min have similarly similar EI and PCI spectra, presumed to be isomers of the same compound.
The setting range of the elemental composition of the original text is wrong. It should be "carbon atom (1-30), hydrogen atom (1-60), nitrogen atom (0-5), oxygen atom (0-5), phosphorus atom (0). -5) and sulfur atom (0-2)".
Figure 7. EI and PCI spectra of unknown compounds with a retention time of 15.17 min in a black ring seal sample (D). The elemental composition of the excimer ion peak and the associated adduct ion in the PCI map is estimated to be C20H20O4.
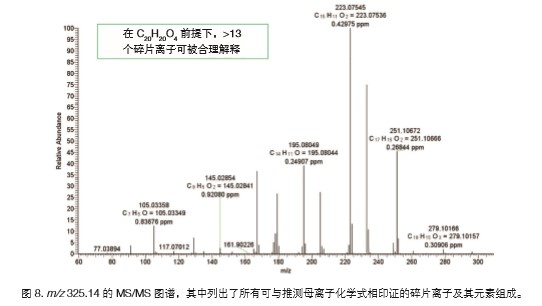
The final step in confirming the elemental composition and structural information of the compound requires reference to the exact mass of the fragment ion. To complete this step, we can directly analyze the fragment ions in the EI spectrum or separately collect the corresponding MS/MS data to confirm that the fragment ions are derived from the parent ion. The ion m/z 325.14 is separated by a quadrupole and introduced into the HCD collision cell to break down. Figure 8 shows the MS/MS spectrum of m/z 325.14. The exact mass of all fragment ions is less than 1 ppm within the tolerance range of the putative parent ion. Thus, even if we have no way to accurately identify the compound, accurate details about the structure of the compound can be extracted.
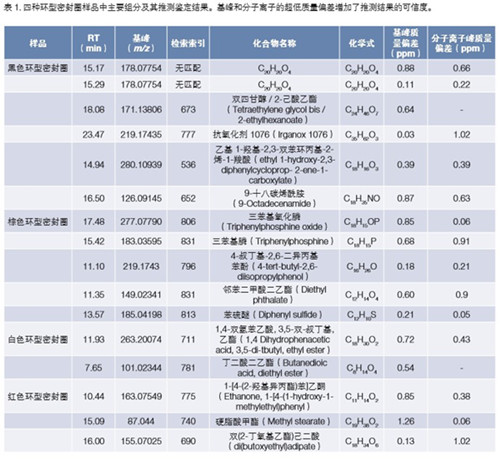
All four ring seal samples were analyzed via the same analytical procedure. The results are summarized in Table 1. We analyzed the highest levels of components in each sample. The results of the identification are based on standard library matching and the exact mass of molecular or adduct ions in the EI and PCI spectra. The black and brown ring seals have the highest levels of compounds in the ethanol extract and the white ring seals. It is apparent from the TIC that the red ring seal sample is contaminated with a cyclosiloxane compound. Contaminants are not listed in the final results list.
in conclusion
The results of this test show that the Q Exactive GC combined quadrupole-electrostatic field orbitrap mass spectrometer can be used in conjunction with TraceFinder software to quickly characterize complex samples and perform structural identification of unknown components. Electrostatic field orbitrap mass spectrometry provides quality information with minimal deviations of all components in a sample, so we can refer to relevant information for rapid and reliable qualitative analysis, regardless of component content.
• Stable, robust chromatographic separation combined with fast data scanning speeds make the Q Exactive GC system an ideal platform for characterizing the chemical composition of complex samples.
• Sub-ppm mass deviation and excellent sensitivity make it possible to reliably identify all chemical components in a sample. The mass resolution of conventional tests is 60,000 FWHM. Combined with the wide dynamic range, it can effectively eliminate the isobaric interference and improve the credibility of compound identification in complex matrices.
• TraceFinder software provides fast, in-depth characterization of ring seal samples and separation and reliable identification of individual components.
• Combine EI and PCI data and compare it to a commercial standard spectral library to infer compound structure. If there is no matching result in the standard library, the elemental composition of the compound can be reliably estimated by the accurate mass information with super high quality accuracy. Based on the exact mass, it is speculated that the identification results can be quickly confirmed or excluded.
references
1. Norwood, DL Understanding the challenges of extractables and leachables for the pharmaceutical industry–safety and regulatory environment for pharmaceuticals. Am. Pharm. Rev., 2007, 10(2), 32–39.
2. Ding, W.; Madsen, G.; Mahajan, E; O'Connor, S; Wong, K. Standardized Extractables Testing Protocol for Single-Use Systems in Biomanufacturing. Pharmaceutical Engineering, 2014, 34(6).
Pet Massager,Cat Massage,Dog Massage,Pet Groomer Tools
Shenzhen Jie Zhong Lian Investment Co., Ltd. , https://www.szmeizon.com
