Screening DNA damage by high-throughput RNA interference technology
-Molecular Devices ImageXpress
At present, the combination of RNAi libraries and high-throughput analysis techniques based on cell levels has broad prospects for us to identify new mechanisms for drug sensitivity and resistance. For example, researchers have performed RNAi screening of whole genomes to identify genes that cause paclitaxel sensitivity in non-small cell lung cancer. This study also confirmed the potential of RNAi technology to find targets that affect cell mitosis in cancer cells.
Molecular Devices has developed a very powerful screening platform, the ImageXpress system. The system has a complete image acquisition and analysis module to quickly generate the desired data.
Here we present how to detect DNA damage by immunofluorescence two-color analysis of phosphorylated histone H2AX and DNA. Phosphorylation of histone H2AX at the serine 139 site is considered to be a sensitive marker of DNA damage caused by drugs or radiation. It is also clarified that we use RNAi to analyze the damage analysis of two-color calibration DNA is a sensitive and rapid automated process, and this method has been proven to effectively find new targets. Figure 1 illustrates the setup method for this analysis.
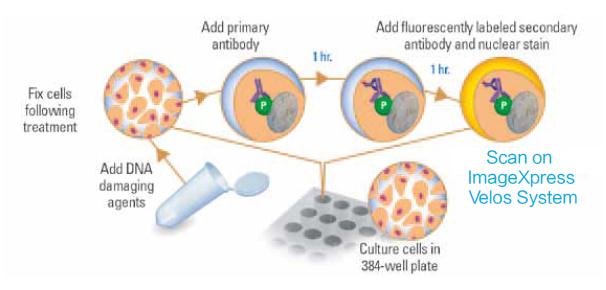
Figure 1 Schematic diagram of DNA damage detection by labeling phosphorylated histone H2AX
Experimental procedure
Cells and their culture conditions. HeLa cells were cultured in DMEM supplemented with 10% FCS. The cells were transferred to a 384-well dark-stained polystyrene plate (Greiner) with a density of about 600 cells per well.
DNA damage was detected using the ImageXpress system. After RNAi and/or DNA damage treatment, the cells were fixed, perforated and the phosphorylation site of serine 139 of histone H2AX was directly immunostained with the corresponding antibody. The secondary antibody used was an anti-rabbit Alexa Fluor-488 (AF-488, Invitrogen), propidium iodide (PI) labeled all nuclei. The ImageXpress laser scanning platform is set to a green filter channel (Ch1, 510-540 nm BP) and a red filter channel (Ch3, 600 nm LP). The full-hole image can be analyzed using a zone-by-region scan of the red channel, and the background is adjusted and corrected by the fluorescence intensity of the integrated green and red channels. In Figure 2 we present images of positive and negative samples.
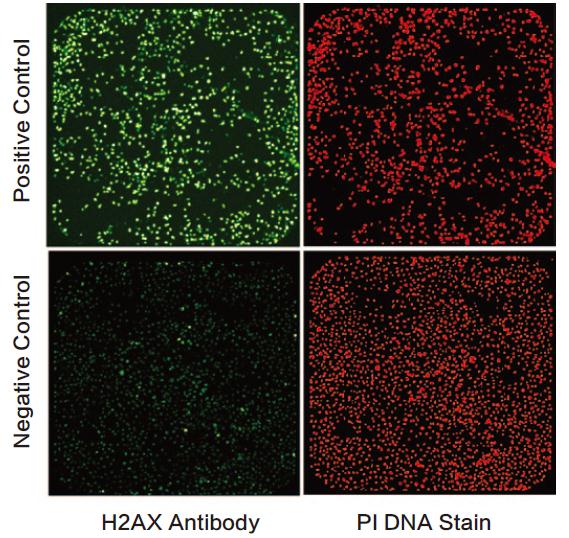
Figure 2 The negative and positive wells of the DNA damage assay were imaged using the ImageXpress system. The cells were grown in 384-well plates with a green channel on the left and a red channel on the right.
Results and discussion
We integrated the ImageXpress system with a mechanical loading arm to screen over 20,000 RNAi (available from the Stanford University Karlene Cimprich Laboratory). Our platform enables automated image processing and quantitative analysis of DNA damage in cells through two-color image processing.
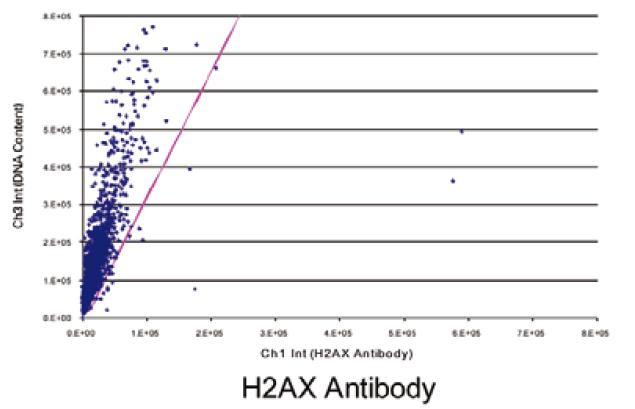
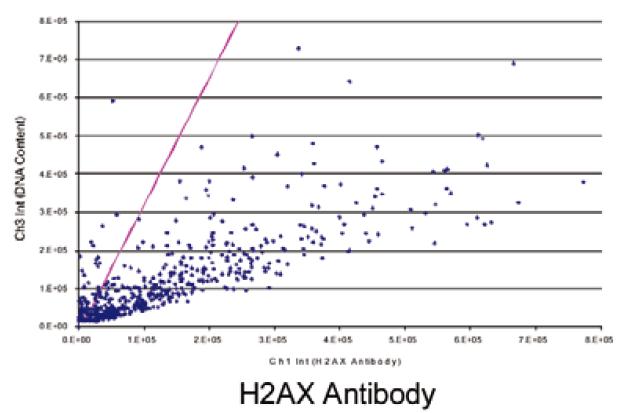
Figure 3 shows a scatter plot of negative (top) and positive (bottom) cell analysis results as shown. The red diagonal line is Ch3/Ch1, indicating the threshold for DNA damage. The negative injury rate was 1.9% and the positive rate was 88.5%.
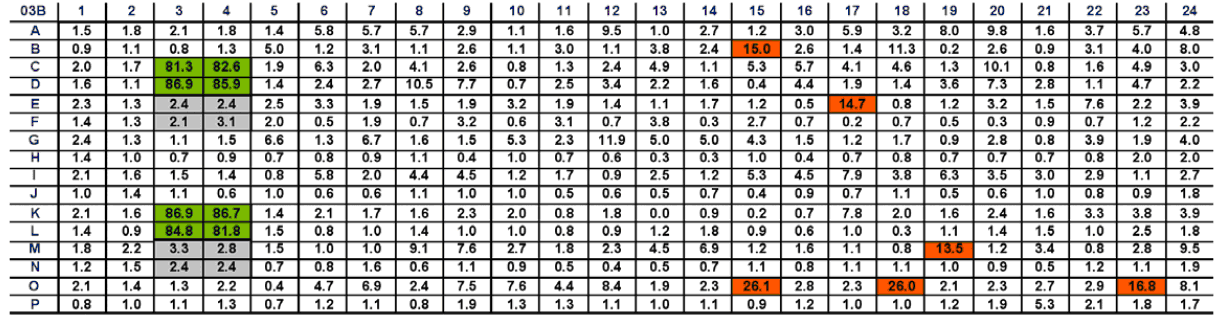
Figure 4 shows the percentage results of RNAi screening for DNA damage arranged in a 384-well plate layout. Negative control empty (8) were marked in gray and positive control wells (8) were marked in green. The Z' value was 0.9, and the damage rate of the other RNAi-treated wells was 13.5%, which was marked orange.
The phosphorylated H2AX and PI stained cells listed in Figure 2 were used as negative and positive controls, and we show their two-parameter bitmap results in Figure 3. The purple dividing line represents the ratio of Ch3/Ch1, and its X-axis intercept is adjustable, and this ratio defines the standard for automated quantitative detection of DNA damage. The ratio of the negative control samples was 1.9% and the ratio of the positive control samples (which had been treated with RNAi technology to increase the probability of DNA damage) was 88.5%.
In Figure 4 we enumerate the percentage of DNA damage in representative 384-well cells. Negative control wells (n=8) are indicated in grey and positive control wells (n=8) are indicated in green. The Z'-prime value is 0.9. The remaining wells treated with RNAi and having a ratio of 13.5% or more were indicated in orange.
in conclusion
Phosphorylation of H2AX has been shown to be an early sensitive marker for detecting ion-irradiated and drug-induced DNA double helix damage. The ImageXpress laser scanning platform we show here can quantify DNA damage by two-color detection. We also illustrate that this two-color DNA damage detection is characterized by high speed, automation, and sensitivity. By designing RNAi, we can screen for genes that induce DNA damage in tumor cells. The platform's unique optical and scanning tools enable simple “plug-in and then-operate†operations, which meet the needs of life science researchers in both academic and industrial fields.
Disposable Gel,Moisturizing Hand Sanitizer,Hand Sanitizer,Hand Sanitizer Dispenser
Wuxi Keni Daily Cosmetics Co.,Ltd , https://www.kenidailycosmetics.com
