Gene chip vs RNA-seq Which is better?
In the last few years, the second-generation sequencing (also known as NGS) is very hot, and the price is getting cheaper. It used to detect the mRNA, miRNA, and LncRNA expressions. It seems that many of them have switched to RNA-seq. So, which is better in the end choose? Answer this question today. One sentence - Â See the purpose of the study.
Common misunderstanding one:
Sequencing is highly accurate and the information obtained is more abundant
Yes, but not right.
First of all, you need to be clear that the concept of detecting and accurately analyzing the amount of gene expression is different. Only when mapping to a certain number of genetic reads can a relatively accurate analysis result be obtained. So how much reliable information RNA-Seq can detect depends entirely on sequencing depth, sequencing depth, and sequencing depth! Unlike chip hybridization, RNA-seq is detected by readings, and many reads (ie, deep sequencing depth) represent a high sampling rate for RNA-seq. The sampling rate is low and the accuracy is naturally low.
So is there an experiment that shows the difference in data accuracy between the chip and the RNA-seq?
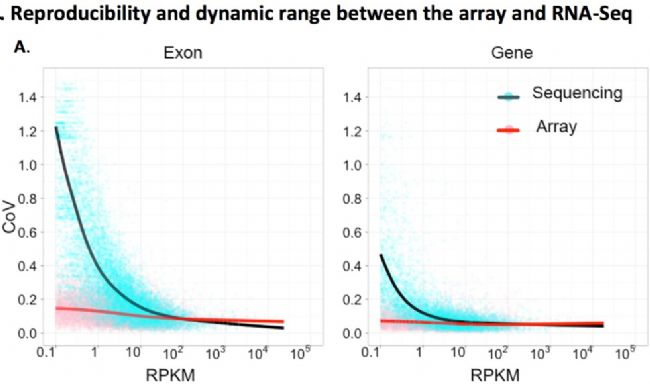
This article published on PNAS will help you make a comparison (PNAS 2011, 108(9): 3707-3712.). The green dot / black line in the figure is the data obtained by sequencing, and the red dot / red line is the data obtained by the chip. In the case of ~50M reads data, when the gene expression abundance is high (when the abscissa RPKM is large), the data quality between the two is very good (the ordinate CoV, that is, the smaller the coefficient of variation, the data The higher the quality, but when the gene expression abundance becomes lower (when the abscissa RPKM is small), the data quality of the RNA-seq drops sharply, while the chip maintains a high level. The conclusion of this article is that ~80% of the genes, RNA-seq data quality / credibility are lower than the chip. The most popular 6G data RNA-seq on the market is actually 40M reads or 20M paired reads, which is almost enough for studying genes with high expression abundance. However, it is not enough for medium and low expression abundance transcripts.
Common Mistakes 2:
RNA-seq can detect both known and unknown genes, and gene chips can only detect known genes, which is a huge limitation.
First, a potential assumption of this view is that some unknown molecules can be found in each sequencing. But for humans, rats, mice, and other model organisms, the genes found have almost been discovered. So whether the gene is known or not is not important in many cases, the focus is on whether the gene is known for its function in the field of your research. Most of the functions of known genes on the chip are still unclear, but it is not advisable to blindly pursue the discovery of new molecules.
RNA-seq is a more appropriate choice in exploratory and non-model biological studies.
Common Mistakes 3:
RNA-seq is now cheaper and much cheaper than gene chips.
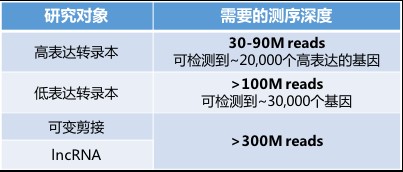
One of the charging standards in sequencing comes from the amount of data (ie, the depth of sequencing). Just now, the most popular RNA-seq service data on the market is 6G/sample, which is 40M reads or 20M paired reads. The chips are cheap. However, if you want to detect medium and low abundance RNA more accurately, you need deeper sequencing to ensure data reliability, which will lead to a sharp increase in sequencing costs. The table below summarizes the sequencing data requirements for some common studies. An article in Nature Biotechnology pointed out that if you want to detect transcripts with very low abundance, such as lncRNA and tran isomers, you need at least 300M reads to achieve 80% data accuracy (Nature biotechnology, 2014, 32). (9): 903-914.).
So what about the chip? Take the Affymetrix HTA series of chips, the amount of data, but the equivalent of 480M reads sequencing depth! Wow, it seems like I saw a lot of money.
Common Misunderstanding 4:
RNA-seq can also detect mutations while measuring expression levels, and gene chips cannot.
Gene chips (here specifically referred to as RNA expression profiles) do not detect mutations. RNA-seq is used to detect RNA abundance by sequencing, and sequence information can be obtained. However, because sequencing itself has an error rate, RNA-seq often has a low sequencing depth, and the obtained mutation information is not accurate. To be accurate, you need a very high depth of sequencing, and then back to the old problem, the cost is basically unacceptable.
So some students have to ask, there are many kinds of gene chips on the market, I don’t know where to start? Xiaobian will talk to you about the choice of gene chip types in the next issue.

Long press and pay attention
Fish Bone Molding 18G100mm,Fish Bone Molding 18G100mm Bolt,Fish Bone Molding 18G100mm Filter,Fish Bone Molding 18G100mm In Inches
Qingdao Beautiful Skin Biotechnology Co., Ltd , https://www.hafilleresthetic.com
